Researchers at the University of Waterloo Introduce Orchid: Revolutionizing Deep Learning with Data-Dependent Convolutions for Scalable Sequence Modeling
In deep learning, especially in NLP, image analysis, and biology, there is an increasing focus on developing models that offer both computational efficiency and robust expressiveness. Attention mechanisms have been revolutionary, allowing for better handling of sequence modeling tasks. However, the computational complexity associated with these mechanisms scales quadratically with sequence length, which becomes a significant bottleneck when managing long-context tasks such as genomics and natural language processing. The ever-increasing need for processing larger and more complex datasets has driven researchers to find more efficient and scalable solutions.
A main challenge in this domain is reducing the computational burden of attention mechanisms while preserving their expressiveness. Many approaches have attempted to address this issue by sparsifying attention matrices or employing low-rank approximations. Techniques such as Reformer, Routing Transformer, and Linformer have been developed to enhance attention mechanisms’ computational efficiency. Yet, these techniques struggle to balance computational complexity and expressive power perfectly. Some models use combinations of these techniques alongside dense attention layers to enhance expressiveness while maintaining computational feasibility.
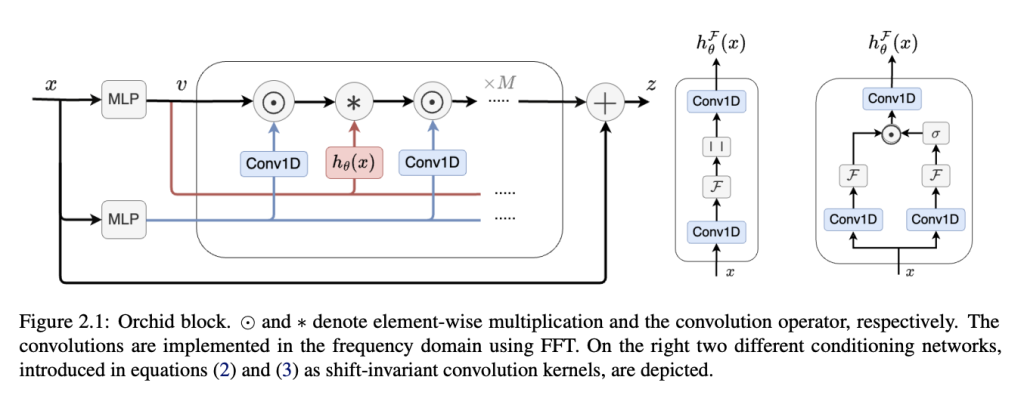
A new architectural innovation known as Orchid has emerged from research at the University of Waterloo. This innovative sequence modeling architecture integrates a data-dependent convolution mechanism to overcome the limitations of traditional attention-based models. Orchid is designed to tackle the inherent challenges of sequence modeling, particularly quadratic complexity. By leveraging a new data-dependent convolution layer, Orchid dynamically adjusts its kernel based on the input data using a conditioning neural network, allowing it to handle sequence lengths up to 131K efficiently. This dynamic convolution ensures efficient filtering of long sequences, achieving scalability with quasi-linear complexity.
The core of Orchid lies in its novel data-dependent convolution layer. This layer adapts its kernel using a conditioning neural network, significantly enhancing Orchid’s ability to filter long sequences effectively. The conditioning network ensures that the kernel adjusts to the input data, strengthening the model’s ability to capture long-range dependencies while maintaining computational efficiency. By incorporating gating operations, the architecture enables high expressivity and quasi-linear scalability with a complexity of O(LlogL). This allows Orchid to handle sequence lengths well beyond the limitations of dense attention layers, demonstrating superior performance in sequence modeling tasks.
The model outperforms traditional attention-based models, such as BERT and Vision Transformers, across domains with smaller model sizes. On the Associative Recall task, Orchid consistently achieved accuracy rates above 99%, with sequences up to 131K. Compared to the BERT-base, the Orchid-BERT-base has 30% fewer parameters yet achieves a 1.0-point improvement in the GLUE score. Similarly, Orchid-BERT-large surpasses BERT-large in GLUE performance while reducing parameter counts by 25%. These performance benchmarks highlight Orchid’s potential as a versatile model for increasingly large and complex datasets.

In conclusion, Orchid successfully addresses the computational complexity limitations of traditional attention mechanisms, offering a transformative approach to sequence modeling in deep learning. Using a data-dependent convolution layer, Orchid effectively adjusts its kernel based on input data, achieving quasi-linear scalability while maintaining high expressiveness. Orchid sets a new benchmark in sequence modeling, enabling more efficient deep-learning models to process ever-larger datasets.
Check out the Paper. All credit for this research goes to the researchers of this project. Also, don’t forget to follow us on Twitter. Join our Telegram Channel, Discord Channel, and LinkedIn Group.
If you like our work, you will love our newsletter..
Don’t Forget to join our 41k+ ML SubReddit
![]()
Nikhil is an intern consultant at Marktechpost. He is pursuing an integrated dual degree in Materials at the Indian Institute of Technology, Kharagpur. Nikhil is an AI/ML enthusiast who is always researching applications in fields like biomaterials and biomedical science. With a strong background in Material Science, he is exploring new advancements and creating opportunities to contribute.












